Abstract
The high occurrence of splicing factor mutations (SFMT) has suggested that RNA splicing dysfunction plays a key pathogenic role in myeloid neoplasia (MN). Nevertheless, transcriptome-wide analysis suggests that MDS is linked with a broad spectrum of splicing lesions also occurring in cases without SFMT. Due to such a vast complexity, traditional approaches for identification of splicing patterns taking into account features in unimodal fashion may not be adequate to dissect the pathophysiologic features in detail, being instead amenable to more complex machine learning (ML) strategies. Indeed, we hypothesize that splicing dysfunction in MDS may be more wide spread and not restricted to SFMT cases consistent with view that MDS is a global spliceopathy.
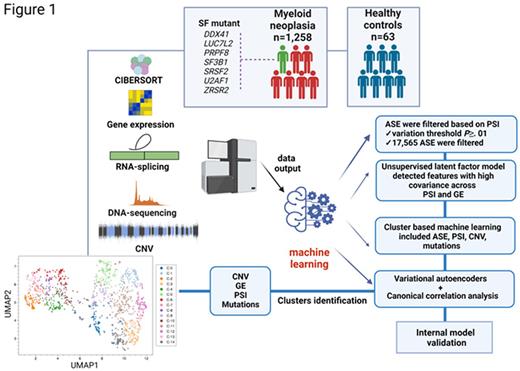
We explore the geno-transcriptomic pattern of MN by merging multi-omics data to build an ML multimodal tool. An integrative analysis of omics data (RNA and WGS of 1258 whole bone marrows of MN patients) along with ML methods coupling variational autoencoders and canonical correlation analysis was applied to identify covarying features including genomic lesions (mutations, deletions, CNV), exon-inclusion ratio (percent spliced in, PSI) and gene expression (GE). CIBERSORT analysis was used to quantify the composition of clonal cells in bulk samples. Patterns of similarities and differences across patients were compared to 63 age-matched healthy controls and an internal validation of the model was also performed (Fig1).
Combination of all features followed by unsupervised clustering identified 15 clusters (C-0 to C-14) resulting in discrete differences in MN subtypes distribution. A distinct definition of the clusters was also seen based on molecular signatures. In our cohort, 54% of patients carried SFMT, further stressing that the mere mutational status of a handful of SFs may not be sufficient to assess the real impact of spliceosome changes. Some clusters were dominated by a high frequency of specific mutations, suggesting a strong functional consequence imprinted by the mutational event. MDS patients were enriched in three clusters with SF3B1MT mainly included in C-1, C-7, C-12. C-1 and C-12 had two distinctive PSI patterns showing decreased splicing efficiency of various genes including DDX20 and KDM3B. SRSF2MT followed similar patterns of distinct PSI in the clusters they populated most (C-8, C-14). C-2 and C-13 contained mainly TET2MT and ASXL1MT. AML patients composed six clusters with two of them (C-9, C-10) consisting of >95% of AML diagnoses. Carriers of DNMT3A, TET2 and FLT3 hits were equally represented in C-0, C-5, C-6 while DNMT3AMT and FLT3MT were equal in C-10. PSI score differences based on ΔPSI (difference in PSI between pairwise clusters) showed high correlation for EZH2 in SRSF2MT, LUC7L2MT and PRPF8MT. PSI similarities were found in subclusters with 1) SF3B1MT, DNMT3AMT, and TET2MT; 2) SF3B1MT with other SFs (PRPF8, DDX41, LUC7L2); 3) TP53MT and DDX41MT possibly suggesting a process of phenocopying and common downstream targets. Using Limma for differential GE analysis we also identified mutational groups clustering together based on GE profiles (group 1: SRSF2, RIT1, JAK2, PTPN11, TET2; group 2: LUC7L2, SF3B1, BCORL1, CSF3R).
Using Ward's criterion based hierarchical clustering, we created a map of defining splicing features per cluster. Combination of our multimodal ML with survival outcome in a continuous fashion by means of survival forests and permutation of latent features identified components with highest associations with OS. Focusing on MDS as the most paradigmatic group inherently characterized by a high heterogeneity, we stratified patients based on low and high risk IPSS-R and found that our ML model was effective in discriminating high risk IPSS-R clusters. Finally, we also found a further refinement in low risk IPSS-R cases, with C-12 (SF3B1MT cluster) showing significant separation from other two close clusters (C-7, C-11).
This is the first study unifying different levels of multi-omics information to examine the genetic picture of MN as a global spliceopathy. The convergence of molecular and transcriptomic pathways might unveil subgroups of seemingly diverse patients sharing common pathogenetic routes and the identification of these pathways might guide rational therapies. To facilitate the use of such integrated approach intertwining PSI, mutations and GE, we created an interactive MutExPSIplot.1
Disclosures
Haferlach:Munich Leukemia Laboratory: Current Employment, Other: Part ownership. Maciejewski:Alexion: Consultancy; Apellis Pharmaceuticals: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal